Chapter 8: Cluster analysis
Figure 1
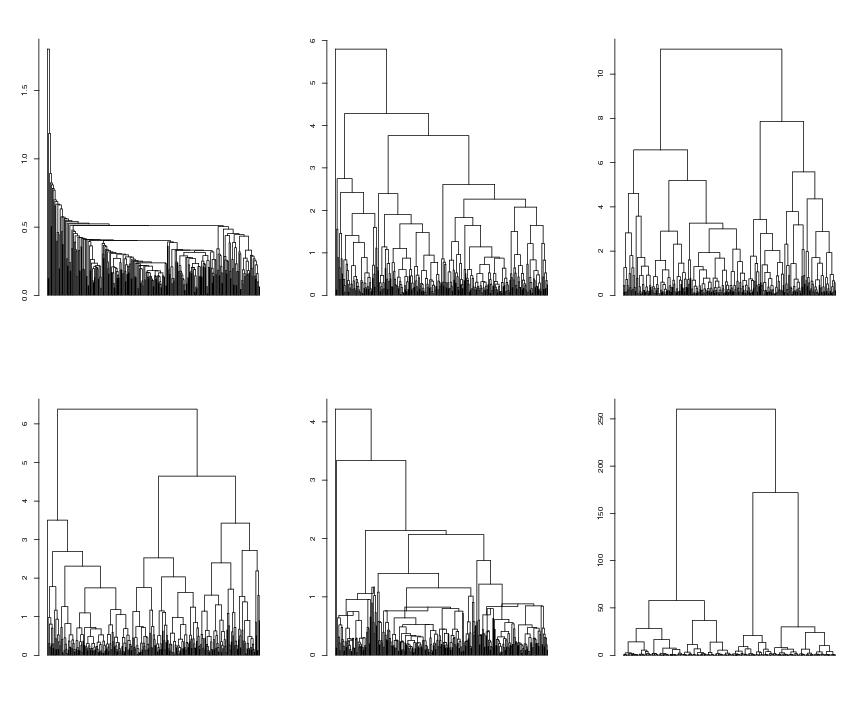
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
dis<-dist(dendat)
method<-"single"
hc1 <- hclust(dis, method=method)
method<-"average"
hc2 <- hclust(dis, method=method)
method<-"complete"
hc3 <- hclust(dis, method=method)
method<-"mcquitty"
hc4 <- hclust(dis, method=method)
method<-"centroid"
hc5 <- hclust(dis, method=method)
method<-"ward"
hc6 <- hclust(dis, method=method)
# frame 1
plot(hc1, hang = -1,xlab="",ylab="",labels=FALSE,main="",sub="")
# frame 2
plot(hc2, hang = -1,xlab="",ylab="",labels=FALSE,main="",sub="")
# frame 3
plot(hc3, hang = -1,xlab="",ylab="",labels=FALSE,main="",sub="")
# frame 4
plot(hc4, hang = -1,xlab="",ylab="",labels=FALSE,main="",sub="")
# frame 5
plot(hc5, hang = -1,xlab="",ylab="",labels=FALSE,main="",sub="")
# frame 6
plot(hc6, hang = -1,xlab="",ylab="",labels=FALSE,main="",sub="")
Figure 2
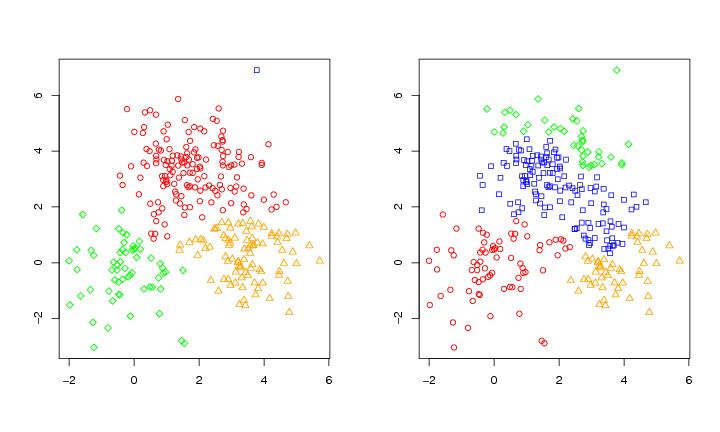
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
dis<-dist(dendat)
method<-"average"
hc2 <- hclust(dis, method=method)
tr2<-dend2parent(hc2,dendat)
method<-"complete"
hc3 <- hclust(dis, method=method)
tr3<-dend2parent(hc3,dendat)
paletti<-c("red","blue","green",
"orange","navy","darkgreen",
"orchid","aquamarine","turquoise",
"pink","violet","magenta","chocolate","cyan",
colors()[50:657],colors()[50:657])
pchvec<-c(21:25)
# frame 1
colothre<-3
col<-colobary.roots(tr2$parent,tr2$level,paletti=paletti,colothre=colothre)
pch<-colobary.roots(tr2$parent,tr2$level,paletti=pchvec,colothre=colothre)
pch<-as.numeric(pch)
plot(dendat,col=col[tr2$pointers],xlab="",ylab="",pch=pch[tr2$pointers])
# frame 2
colothre<-6
col<-colobary.roots(tr3$parent,tr3$level,paletti=paletti,colothre=colothre)
pch<-colobary.roots(tr3$parent,tr3$level,paletti=pchvec,colothre=colothre)
pch<-as.numeric(pch)
plot(dendat,col=col[tr3$pointers],xlab="",ylab="",pch=pch[tr3$pointers])
Figure 3
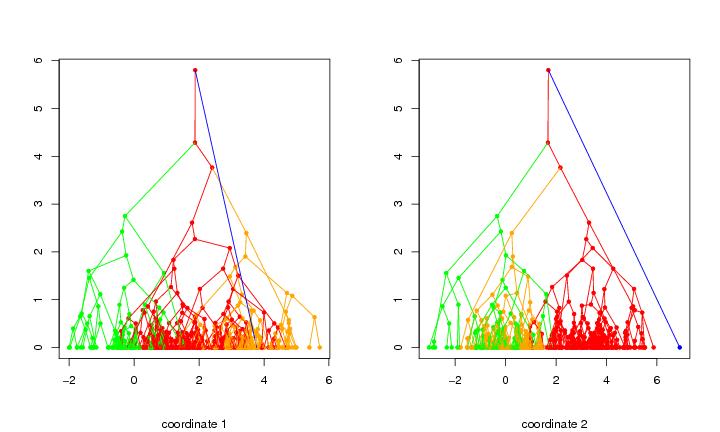
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
dis<-dist(dendat)
method<-"average"
hc2 <- hclust(dis, method=method)
tr2<-dend2parent(hc2,dendat)
# frame 1
plotbary(tr2,modelabel=F,coordi=1,colometh="cluster",colothre=3)
# frame 2
plotbary(tr2,modelabel=F,coordi=2,colometh="cluster",colothre=3)
Figure 4
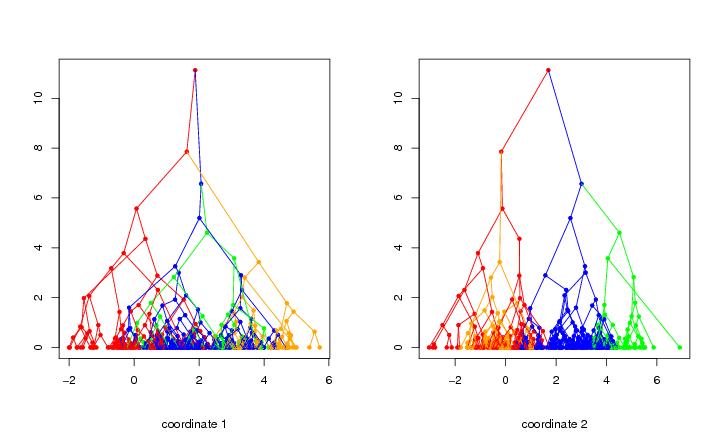
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
dis<-dist(dendat)
method<-"complete"
hc3 <- hclust(dis, method=method)
tr3<-dend2parent(hc3,dendat)
# frame 1
plotbary(tr3,modelabel=FALSE,coordi=1,colometh="cluster",colothre=6)
# frame 2
plotbary(tr3,modelabel=FALSE,coordi=2,colometh="cluster",colothre=6)
Figure 5
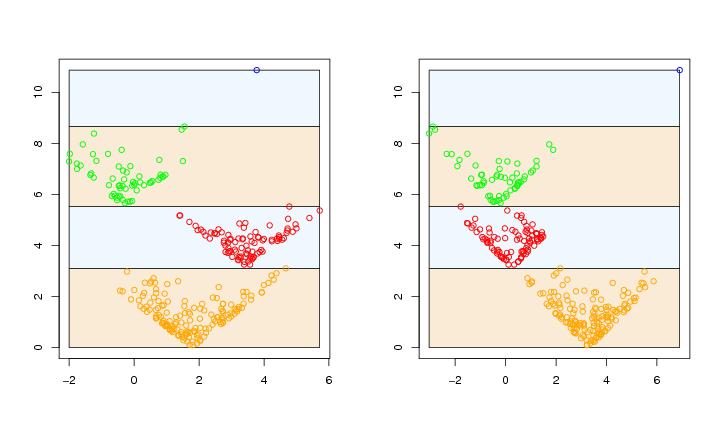
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
algo<-"average"
k<-4
levelmethod<-"center"
range<-"local"
paletti<-c("orange","red","green","blue")
# frame 1
paraclus(dendat,algo=algo,k=k,paletti=paletti,
range=range,levelmethod=levelmethod,terminal=FALSE,coordi=1)
# frame 2
paraclus(dendat,algo=algo,k=k,method=method,paletti=paletti,
range=range,levelmethod=levelmethod,terminal=FALSE,coordi=2)
Figure 6
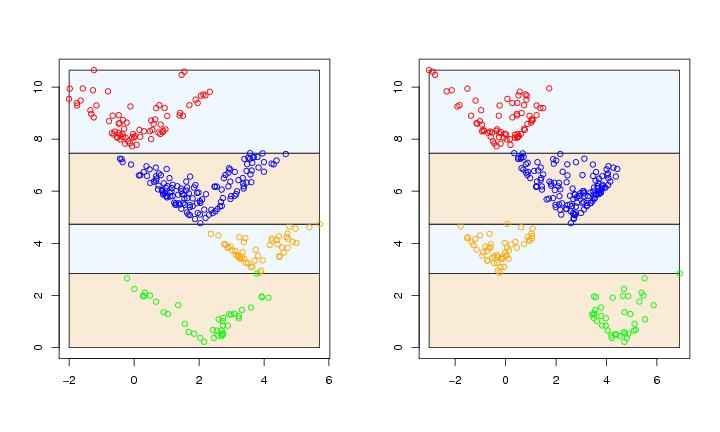
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
algo<-"complete" #"hclust" #"kmeans"
k<-4
levelmethod<-"center" #"random"
range<-"local" #"other"
paletti<-c("green","orange","blue","red")
# frame 1
paraclus(dendat,algo=algo,k=k,method=method,scatter=FALSE,paletti=paletti,
range=range,levelmethod=levelmethod,terminal=FALSE,coordi=1)
# frame 2
paraclus(dendat,algo=algo,k=k,method=method,scatter=FALSE,paletti=paletti,
range=range,levelmethod=levelmethod,terminal=FALSE,coordi=2)
Figure 7
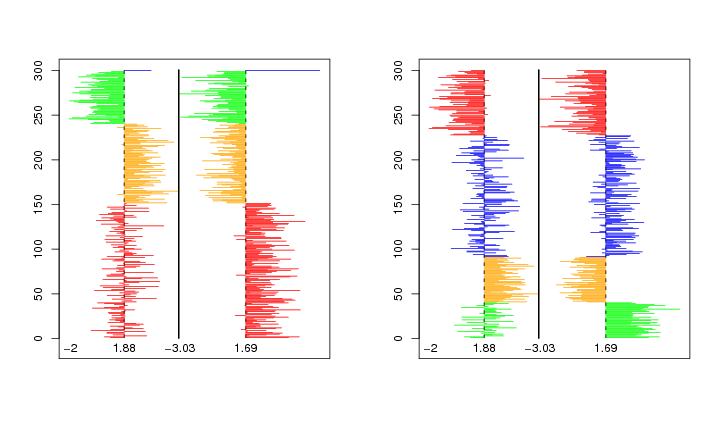
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
dis<-dist(dendat)
method<-"average"
hc2 <- hclust(dis, method=method)
method<-"complete"
hc3 <- hclust(dis, method=method)
# frame 1
luokat=cutree(hc2,k=4)
pale<-c("red","orange","green","blue")
col<-pale[luokat]
permu<-order(luokat)
graph.matrix(dendat,permu=permu,col=col,ystart=-10)
# frame 2
luokat=cutree(hc3,k=4)
pale<-c("green","orange","blue","red")
col<-pale[luokat]
permu<-order(luokat)
graph.matrix(dendat,permu=permu,col=col,ystart=-10)
Figure 8
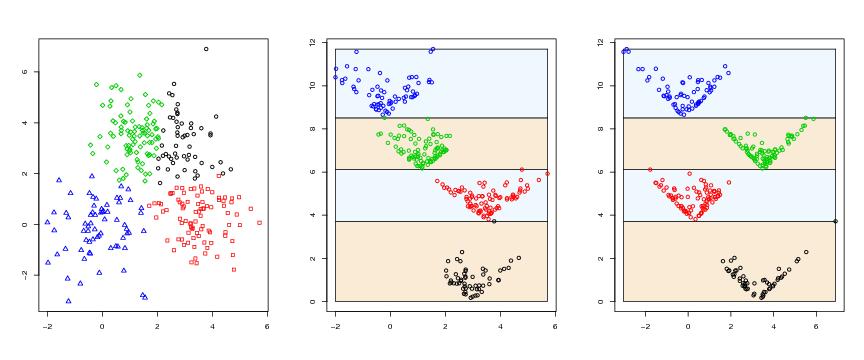
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
k<-4
startind<-c(1:k)
starters<-dendat[startind,]
cl<-kmeans(dendat,k,centers=starters)
ct<-cl$cluster
algo<-"kmeans" #"hclust"
levelmethod<-"center" #"random"
range<-"local" #"other"
pchvec<-c(21:25)
# frame 1
plot(dendat,col=ct,xlab="",ylab="",pch=pchvec[ct])
# frame 2
paraclus(dendat,algo=algo,k=k,
range=range,levelmethod=levelmethod,terminal=FALSE,coordi=1)
# frame 3
paraclus(dendat,algo=algo,k=k,method=method,
range=range,levelmethod=levelmethod,terminal=FALSE,coordi=2)
Figure 9
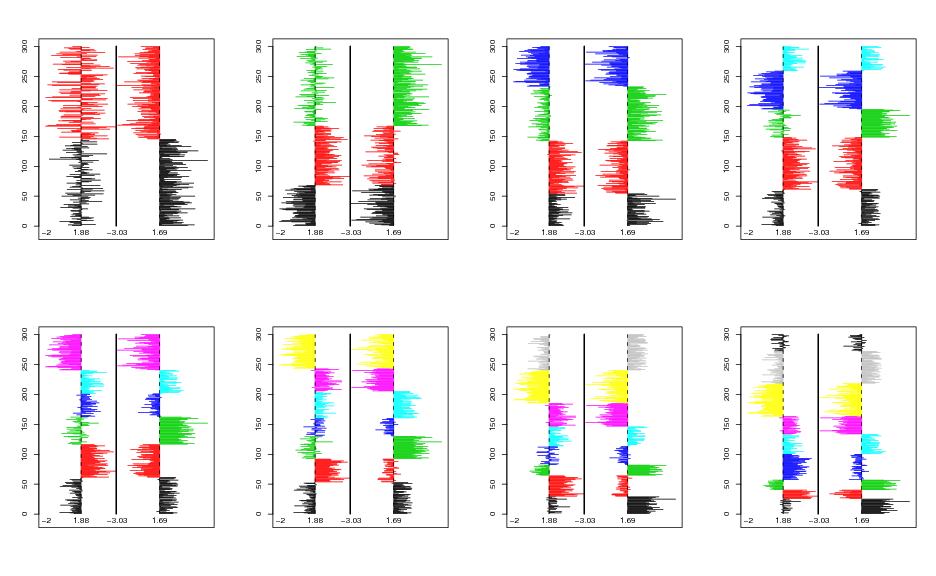
siemen<-5
n<-300
dendat<-sim.data(n=n,seed=siemen,type="mulmod")
# frames 1-4
for (i in 2:5){
k<-i
starters<-dendat[c(1:k),]
cl<-kmeans(dendat,k,centers=starters)
luokat<-cl$cluster
permu<-order(luokat)
graph.matrix(dendat,permu=permu,col=luokat,ystart=-10)
}
# frames 5-8
for (i in 6:9){
k<-i
starters<-dendat[c(1:k),]
cl<-kmeans(dendat,k,centers=starters)
luokat<-cl$cluster
permu<-order(luokat)
graph.matrix(dendat,permu=permu,col=luokat,ystart=-10)
}
Figure 10

n<-700
dendat<-sim.data(n=n,type="nested",seed=1)
N<-c(40,40)
pcf<-sim.data(N=N,type="nested")
dp<-draw.pcf(pcf,pnum=N)
# frame 1
plot(dendat,xlab="",ylab="")
# frame 2
contour(dp$x,dp$y,dp$z,drawlabels=FALSE,nlevels=20)
Figure 11
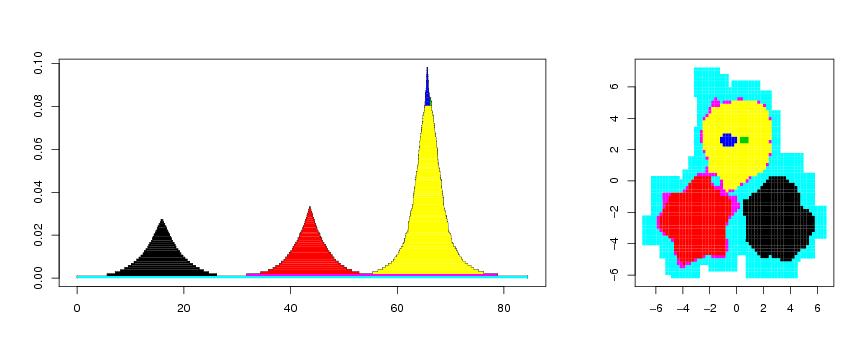
n<-700
dendat<-sim.data(n=n,type="nested",seed=1)
N<-c(64,64)
kernel<-"epane"
ke<-pcf.kern(dendat,h=1,N=N,kernel=kernel)
dp<-draw.pcf(ke,pnum=N)
lst<-leafsfirst(ke)
lst2<-treedisc(lst,ke,ngrid=100)
col<-colobary(lst$parent,paletti=seq(1:2000))
kaanto<-lst$infopointer
for (i in 1:length(kaanto)) kaanto[lst$infopointer[i]]<-i
mat<-matrix(c(1:2),1,2)
layout(mat,widths=c(2,1))
# frame 1
plotvolu(lst2,colo=TRUE,paletti=seq(1:2000))
# frame 2
draw.levset(ke,propor=0,col=col[kaanto])
Figure 12
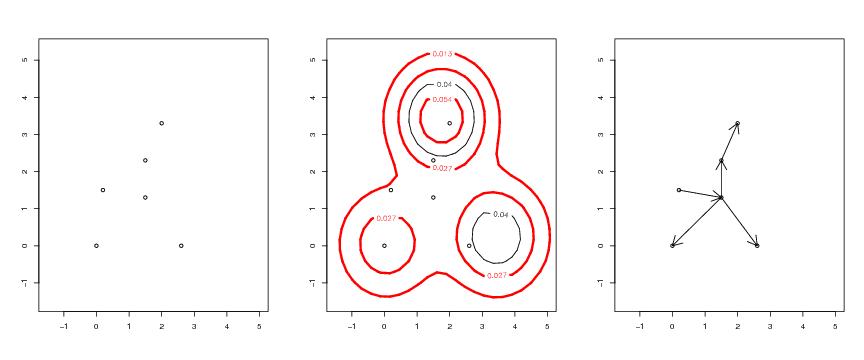
N<-c(32,32)
eg<-sim.mulmod(N=N)
lf<-leafsfirst(eg)
ngrid<-4
lf.redu<-treedisc(lf,eg,ngrid=ngrid)
lf.plot<-lf.redu
lf.plot$level<-c(0,1,1,1,2,2,,3)
stepsi<-lf$maxdis/(ngrid+1)
rad<-seq(stepsi,lf$maxdis-stepsi,stepsi)
roundrad<-round(rad,digits=3)
dm<-draw.pcf(eg)
d<-2
n<-6
dendat<-matrix(0,n,d)
dendat[1,]<-c(0.2,1.5)
dendat[2,]<-c(1.5,1.3)
dendat[3,]<-c(0,0)
dendat[4,]<-c(2.6,0)
dendat[5,]<-c(1.5,2.3)
dendat[6,]<-c(2,3.3)
xala<--1.5
xyla<-5
yala<--1.5
yyla<-5.3
# frame 1
plot(dendat,xlab="",ylab="",xlim=c(xala,xyla),ylim=c(yala,yyla))
# frame 2
plot(dendat,xlab="",ylab="",xlim=c(xala,xyla),ylim=c(yala,yyla))
contour(dm$x,dm$y,dm$z,levels=roundrad,add=TRUE,
col=c("red","red","black","red"),lwd=c(3,3,1,3))
# frame 3
plot(dendat,xlab="",ylab="",xlim=c(xala,xyla),ylim=c(yala,yyla))
arrows(dendat[1,1],dendat[1,2],dendat[2,1],dendat[2,2],length=0.15)
arrows(dendat[2,1],dendat[2,2],dendat[3,1],dendat[3,2],length=0.15)
arrows(dendat[2,1],dendat[2,2],dendat[4,1],dendat[4,2],length=0.15)
arrows(dendat[2,1],dendat[2,2],dendat[5,1],dendat[5,2],length=0.15)
arrows(dendat[5,1],dendat[5,2],dendat[6,1],dendat[6,2],length=0.15)
Figure 13
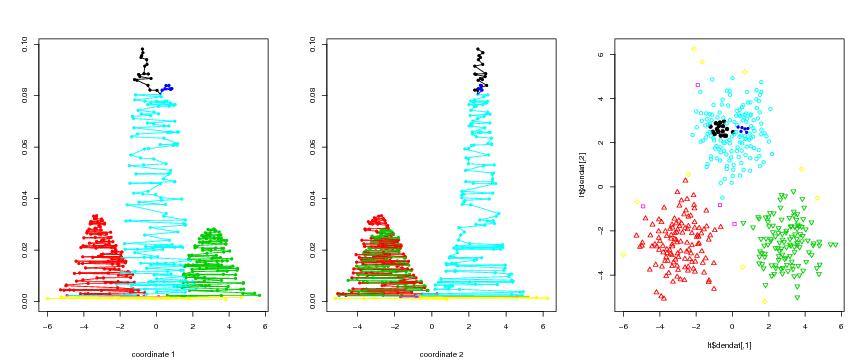
n<-700
dendat<-sim.data(n=n,type="nested",seed=1)
N<-c(64,64)
kernel<-"epane"
ke<-pcf.kern(dendat,h=1,N=N,kernel=kernel)
dp<-draw.pcf(ke,pnum=N)
lst<-leafsfirst(ke)
lt<-liketree(dendat,ke,lst)
# frame 1
plotbary(lt,paletti=seq(1:2000),coordi=1) #,lines=FALSE)
# frame 2
plotbary(lt,paletti=seq(1:2000),coordi=2)
# frame 3
paletti<-seq(1:2000)
col<-colobary(lt$parent,paletti)
pchvec=c(19,24,25,20,21,22,23)
plot(lt$dendat,col=col,pch=pchvec[col])
Figure 14
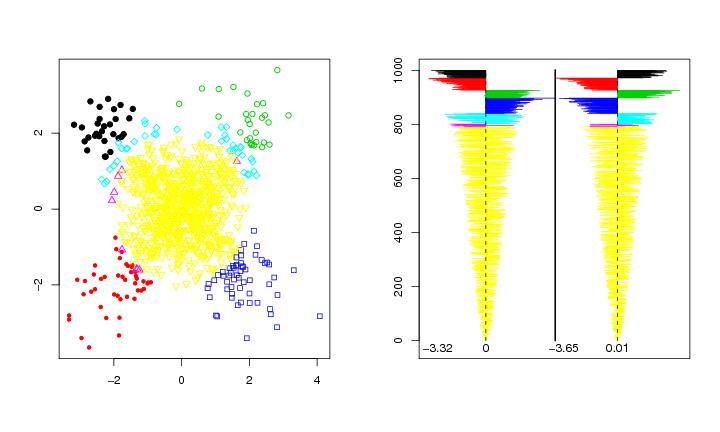
n<-1000
dendat<-sim.data(n=n,type="cross",seed=1)
rho<-0.65
tt<-leafsfirst(dendat=dendat,rho=rho)
colo<-tree.segme(tt,paletti=seq(1,1000))
# frame 1
pchvec<-c(19:25)
plot(dendat,col=colo,pch=pchvec[colo],xlab="",ylab="")
# frame 2
graph.matrix(dendat,tt=tt,ystart=-25,shift=0.5)
Figure 15
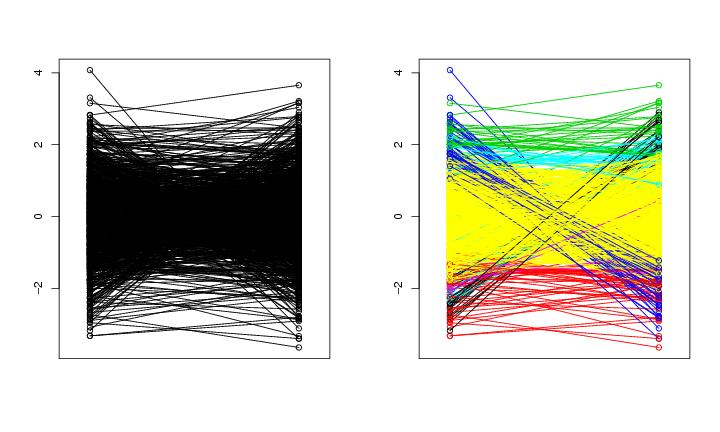
n<-1000
dendat<-sim.data(n=n,type="cross",seed=1)
rho<-0.65
tt<-leafsfirst(dendat=dendat,rho=rho)
colo<-tree.segme(tt)
# frame 1
paracoor(dendat)
# frame 2
paracoor(dendat,paletti=colo)
