Chapter 18: Multivariate adaptive histograms
Figure 1
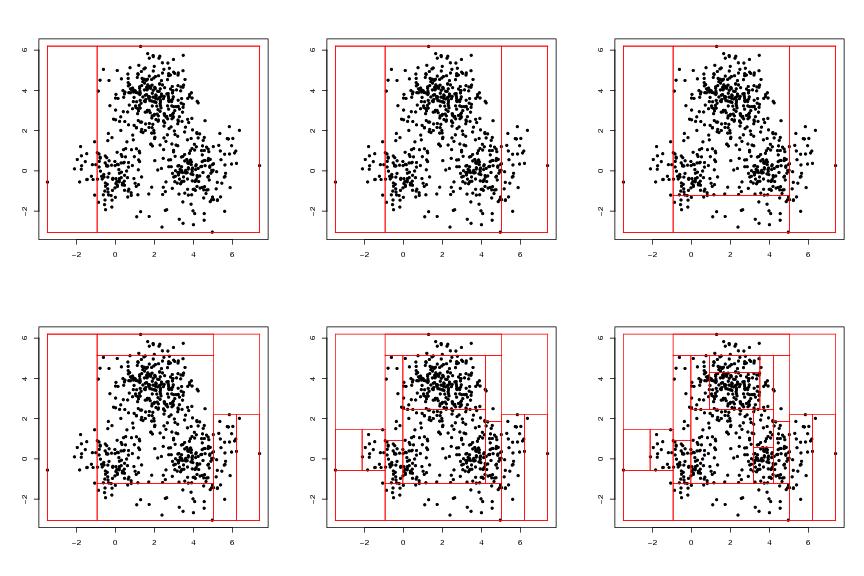
n<-600
type<-"mulmodII"
seed<-5
dendat<-sim.data(n=n,type=type,seed=seed)
leafs<-c(2,3,4,7,15,20)
# frames 1-6
for (i in 1:length(leafs)){
leaf<-leafs[i]
tr<-eval.greedy(dendat,leaf=leaf)
pa<-partition(tr) # pa<-partigen(bt)#,grid=FALSE)
plotparti(pa,dendat=dendat,pch=20,col="red")
}
Figure 2
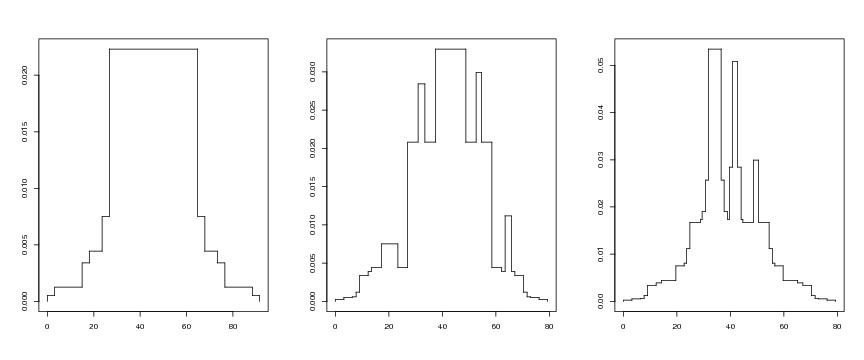
n<-600
type<-"mulmodII"
seed<-5
dendat<-sim.data(n=n,type=type,seed=seed)
maxleaf<-20
lg<-lstseq.greedy(dendat,maxleaf,lstree=TRUE)
sele<-c(14,6,1)
# frame 1
j<-1
i<-sele[j]
lst<-lg$lstseq[[i]]
plotvolu(lst)
# frame 2
j<-2
i<-sele[j]
lst<-lg$lstseq[[i]]
plotvolu(lst)
# frame 3
j<-3
i<-sele[j]
lst<-lg$lstseq[[i]]
plotvolu(lst)
Figure 3
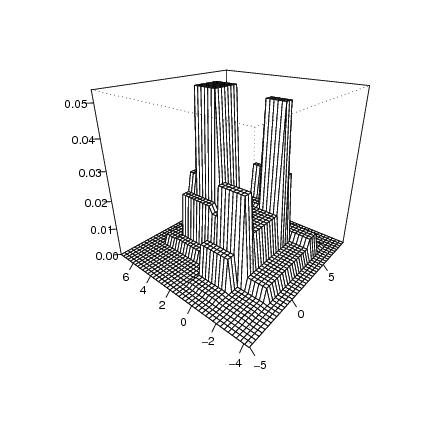
n<-600
type<-"mulmodII"
seed<-5
dendat<-sim.data(n=n,type=type,seed=seed)
maxleaf<-20
lg<-lstseq.greedy(dendat,maxleaf) #,algo=TRUE)
# frame
pcf<-lg$pcfseq[[1]]
dp<-draw.pcf(pcf)
persp(dp$x,dp$y,dp$z,phi=25,theta=-50,col="white",ticktype="detailed",
xlab="",ylab="",zlab="")
Figure 4
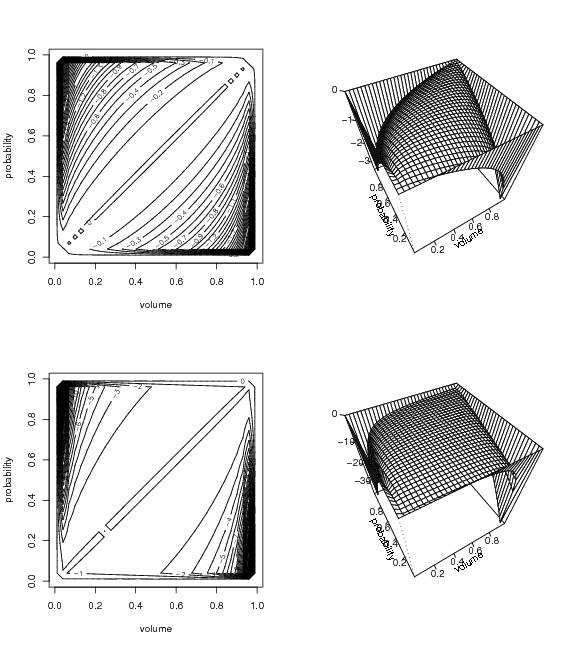
# note: you need to install programs in denpro.R
splitsele<-function(contrast="loglik",vmax=1,pmax=1,
support=c(0.01,vmax-0.01,0.01,0.99),N=c(32,32))
{
d<-length(N)
recnum<-prod(N)
value<-matrix(0,recnum,1)
index<-matrix(0,recnum,d)
lowsuppo<-matrix(0,d,1)
for (i in 1:d) lowsuppo[i]<-support[2*i-1]
step<-matrix(0,d,1)
for (i in 1:d) step[i]<-(support[2*i]-support[2*i-1])/N[i]
for (i in 1:recnum){
inde<-digit(i-1,N)+1
point<-lowsuppo+step*inde-step/2
v<-point[1]
p<-point[2]
if (contrast=="loglik")
valli<--p*log(p/v)-(pmax-p)*log((pmax-p)/(vmax-v))
else
valli<--p^2/v-(pmax-p)^2/(vmax-v)
value[i]<-valli
index[i,]<-inde
}
down<-index-1
high<-index
pcf<-list(
value=value,index=index,
down=down,high=high,
support=support,N=N)
return(pcf)
}
# frame 1
N<-c(70,70)
contrast<-"loglik"
pcf<-splitsele(contrast=contrast)
dp<-draw.pcf(pcf,pnum=N)
contour(dp$x,dp$y,dp$z,nlevels=40,xlab="volume",ylab="probability")
# frame 2
N<-c(50,50)
persp(dp$x,dp$y,dp$z,phi=50,theta=-30,
xlab="volume",ylab="probability",zlab="",ticktype="detailed")
# frame 3
N<-c(70,70)
contrast<-"l2"
pcf<-splitsele(contrast=contrast)
dp<-draw.pcf(pcf,pnum=N)
contour(dp$x,dp$y,dp$z,nlevels=40,xlab="volume",ylab="probability")
# frame 4
N<-c(50,50)
persp(dp$x,dp$y,dp$z,phi=50,theta=-30,
xlab="volume",ylab="probability",zlab="",ticktype="detailed")
Figure 5
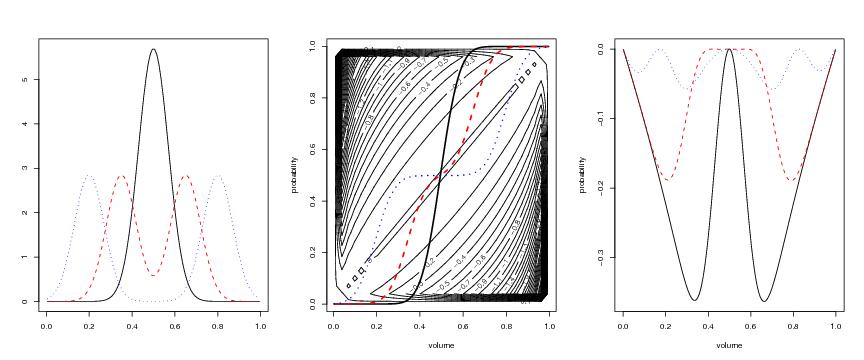
splitsele<-function(contrast="loglik",vmax=1,pmax=1,
support=c(0.01,vmax-0.01,0.01,0.99),N=c(32,32))
{
d<-length(N)
recnum<-prod(N)
value<-matrix(0,recnum,1)
index<-matrix(0,recnum,d)
lowsuppo<-matrix(0,d,1)
for (i in 1:d) lowsuppo[i]<-support[2*i-1]
step<-matrix(0,d,1)
for (i in 1:d) step[i]<-(support[2*i]-support[2*i-1])/N[i]
for (i in 1:recnum){
inde<-digit(i-1,N)+1
point<-lowsuppo+step*inde-step/2
v<-point[1]
p<-point[2]
if (contrast=="loglik")
valli<--p*log(p/v)-(pmax-p)*log((pmax-p)/(vmax-v))
else
valli<--p^2/v-(pmax-p)^2/(vmax-v)
value[i]<-valli
index[i,]<-inde
}
down<-index-1
high<-index
pcf<-list(
value=value,index=index,
down=down,high=high,
support=support,N=N)
return(pcf)
}
splitsele.slice<-function(vmax=1,pmax=1,
step=0.01,vvec=seq(step,vmax-step,step),type="1d2modal",
contrast="loglik",diff=0.1,sig1=0.02)
{
y<-matrix(0,length(vvec),1)
sd<-sim.data(N=length(vvec),distr=TRUE,type=type,diff=diff,sig1=sig1)
pvec<-matrix(0,length(vvec),1)
for (i in 1:length(vvec)) pvec[i]<-sd$value[i]
for (i in 1:length(vvec)){
v<-vvec[i]
p<-pvec[i]
if (contrast=="loglik")
valli<--p*log(p/v)-(pmax-p)*log((pmax-p)/(vmax-v))
else
valli<--p^2/v-(pmax-p)^2/(vmax-v)
y[i]<-valli
}
return(list(x=vvec,y=y))
}
# frame 1
type<-"diff1d"
sig<-0.07
diffet<-c(0,0.15,0.3)
colot<-c(1,2,4,3)
lty<-c(1,2,3,4)
N<-100
sdd<-sim.data(N=N,type=type,diff=diffet[1],sig1=sig)
dpsdd<-draw.pcf(sdd,pnum=60)
plot(dpsdd$x,dpsdd$y,type="l",col=colot[1],xlab="",ylab="")
for (i in 2:3){
diff<-diffet[i]
sdd<-sim.data(N=N,type=type,diff=diff,sig1=sig)
dpsdd<-draw.pcf(sdd,pnum=60)
matplot(dpsdd$x,dpsdd$y,type="l",add=TRUE,col=colot[i],lty=lty[i])
}
# frame 2
contrast<-"loglik" #"l2" #"loglik"
pcf<-splitsele(contrast=contrast)
N<-c(100,100)
dp<-draw.pcf(pcf,pnum=N)
contour(dp$x,dp$y,dp$z,nlevels=40,xlab="volume",ylab="probability")
for (i in 1:3){
diff<-diffet[i]
sd<-sim.data(N=100,type=type,diff=diff,distr=TRUE,sig1=sig)
dpsd<-draw.pcf(sd,pnum=60)
points(dpsd$x,dpsd$y,type="l",col=colot[i],lwd=2,lty=lty[i])
}
# frame 3
step<-0.001
sp<-splitsele.slice(contrast=contrast,type=type,diff=diffet[1],step=step,
sig1=sig)
plot(sp$x,sp$y,xlab="volume",ylab="probability",type="l",col=colot[1])
for (i in 2:3){
diff<-diffet[i]
sp<-splitsele.slice(contrast=contrast,type=type,diff=diff,step=step,
sig1=sig)
matplot(sp$x,sp$y,xlab="volume",ylab="probability",type="l",col=colot[i],
add=TRUE,lty=lty[i])
}
Figure 6
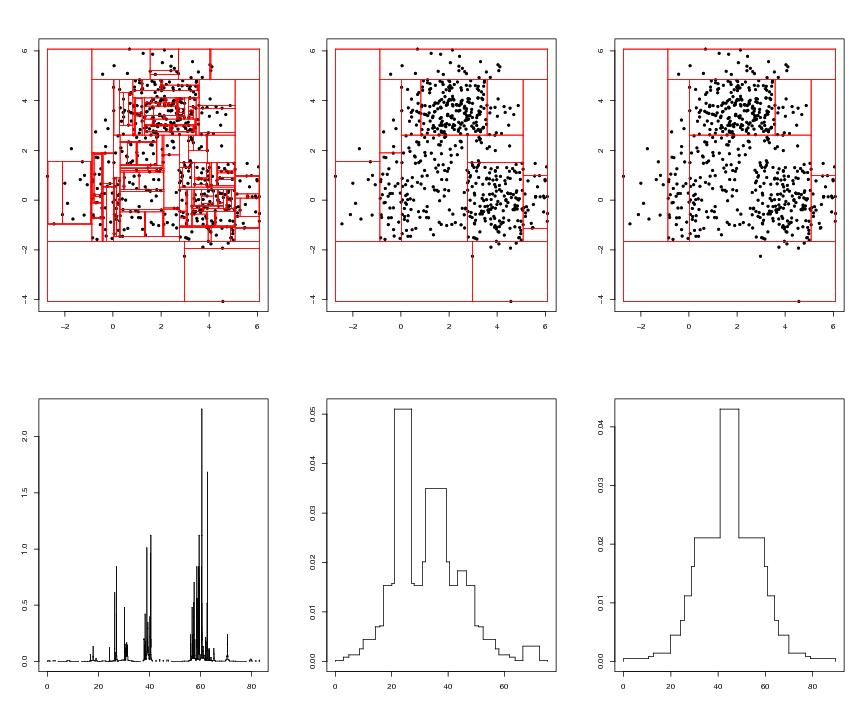
n<-600
type<-"mulmodII"
seed<-2
dendat<-sim.data(n=n,type=type,seed=seed)
minlkm<-5
bt<-densplit(dendat,minobs=minlkm)
treeseq<-prune(bt)
treeseq$leafs
# frame 1
leafnum<-treeseq$leafs[1]
eva1<-eval.pick(treeseq,leafnum)
pa<-partition(eva1,zerorecs=TRUE)
plotparti(pa,dendat=dendat,pch=20,col="red")
# frame 2
leafnum<-16
eva2<-eval.cart(dendat,leafnum,minobs=minlkm)
pa<-partition(eva2,zerorecs=TRUE)
plotparti(pa,dendat=dendat,pch=20,col="red")
# frame 3
leafnum<-10
eva3<-eval.cart(dendat,leafnum,minobs=minlkm)
pa<-partition(eva3,zerorecs=TRUE)
plotparti(pa,dendat=dendat,pch=20,col="red")
# frame 4
lst<-leafsfirst(eva1)
plotvolu(lst)
# frame 5
lst<-leafsfirst(eva2)
plotvolu(lst)
# frame 6
lst<-leafsfirst(eva3)
plotvolu(lst)
Figure 7
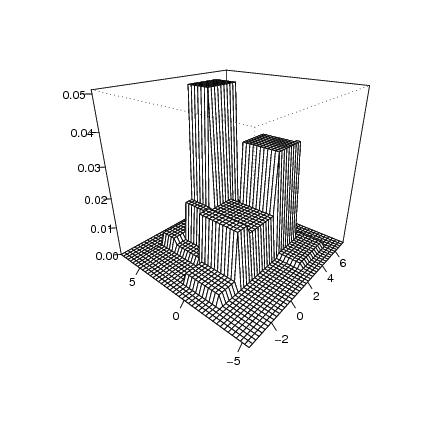
n<-600
type<-"mulmodII"
seed<-2
dendat<-sim.data(n=n,type=type,seed=seed)
minlkm<-5
leafnum<-16
eva2<-eval.cart(dendat,leafnum,minobs=minlkm)
# frame
dp<-draw.pcf(eva2)
persp(dp$x,dp$y,dp$z,phi=25,theta=-50,col="white",ticktype="detailed",
xlab="",ylab="",zlab="")
Figure 8
dendat<-sim.data(n=600,seed=5,type="mulmodII")
leaf<-15
seed<-1
sample="baggworpl"
prune="on"
B<-50
eva<-eval.bagg(dendat,B,leaf,seed=seed,sample=sample,prune=prune)
dp<-draw.pcf(eva,pnum=c(60,60))
lst<-leafsfirst(eva)
lst2<-treedisc(lst,eva,ngrid=100)
# frame 1
persp(dp$x,dp$y,dp$z,phi=25,theta=-50, #phi=25,theta=100,
ticktype="detailed",xlab="",ylab="",zlab="")
# frame 2
plotvolu(lst2)
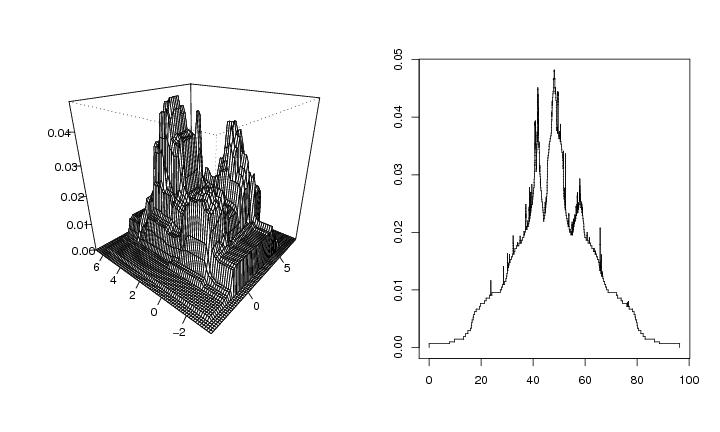
Figure 9
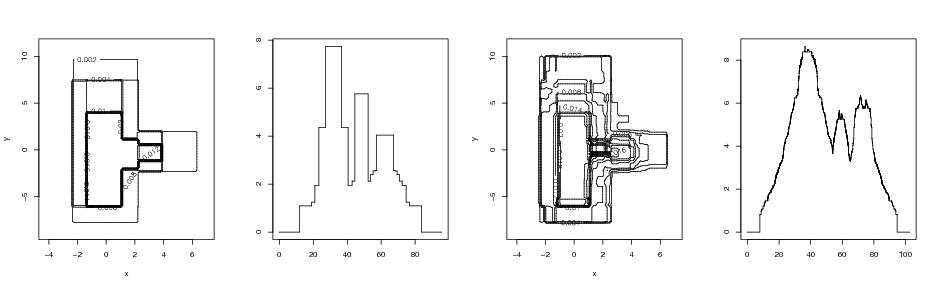
n<-1000
seed<-5
dendat<-sim.data(n=n,type="fox",seed=seed)
leaf<-15
seed<-5
sample="baggworpl"
prune="on"
B<-20
eva<-eval.bagg(dendat,B,leaf,seed=seed,sample=sample,prune=prune)
dp<-draw.pcf(eva,pnum=c(60,60))
lev<-0.002
refe<-c(0,0)
st<-leafsfirst(eva,lev=lev,refe=refe)
minlkm<-5
bt<-densplit(dendat,minobs=minlkm)
treeseq<-prune(bt)
treeseq$leafs
leafnum<-15
eva1<-eval.pick(treeseq,leafnum)
dp1<-draw.pcf(eva1,pnum=c(60,60))
st2<-leafsfirst(eva1,lev=0.002,refe=refe)
# frame 1
contour(dp1$x,dp1$y,dp1$z,xlab="x",ylab="y",nlevels=10)
# frame 2
plotvolu(st2)
# frame 3
contour(dp$x,dp$y,dp$z,xlab="x",ylab="y",nlevels=15)
# frame 4
plotvolu(st)
