Chapter 7: Scales of density estimates
Figure 1
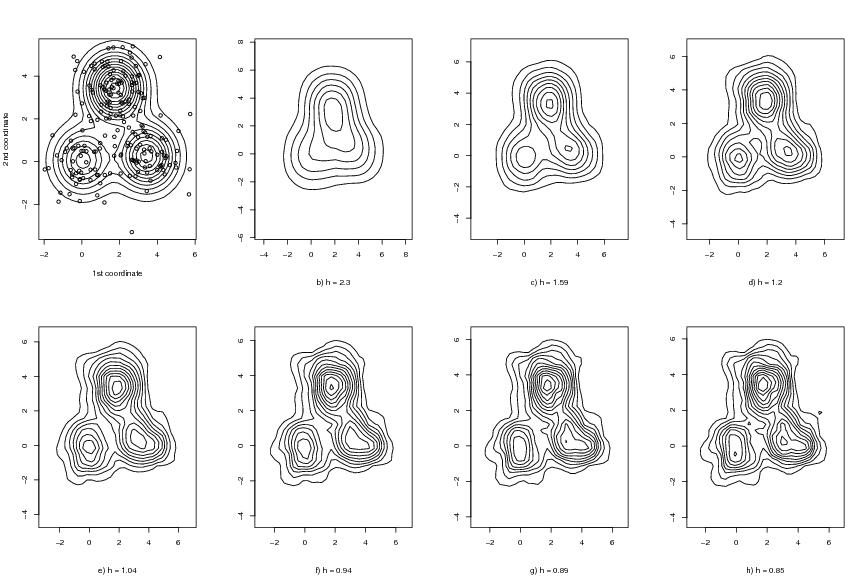
dendat<-sim.data(n=200,seed=5,type="mulmod")
h1<-0.85
h2<-2.3
lkm<-100
base<-10
hseq<-hgrid(h1,h2,lkm,base)
N<-c(30,30)
# Warning: the calculation can take a long time
estiseq<-lstseq.kern(dendat,hseq,N,lstree=TRUE,level=0.1,kernel="epane")
egf<-sim.data(N=N,type="mulmod")
dpf<-draw.pcf(egf)
hind<-c(1,20,40,55,70,85,100)
for (i in 1:7){
pcf<-estiseq$pcfseq[[hind[i]]]
if (i==1) pcflist<-list(pcf)
else pcflist<-c(pcflist,list(pcf))
}
lapeli<-c("b)","c)","d)","e)","f)","g)","h)")
# frame 1
plot(dendat,xlab="1st coordinate",ylab="2nd coordinate")
contour(dpf$x,dpf$y,dpf$z,drawlabels=FALSE,add=TRUE)
# frames 2-8
for (i in 1:length(hind)){
pcf<-pcflist[[i]]
dp<-draw.pcf(pcf)
contour(dp$x,dp$y,dp$z,drawlabels=FALSE)
title(sub=paste(lapeli[i],"h =",
as.character(round(estiseq$hseq[hind[i]],digits=2))))
}
Figure 2
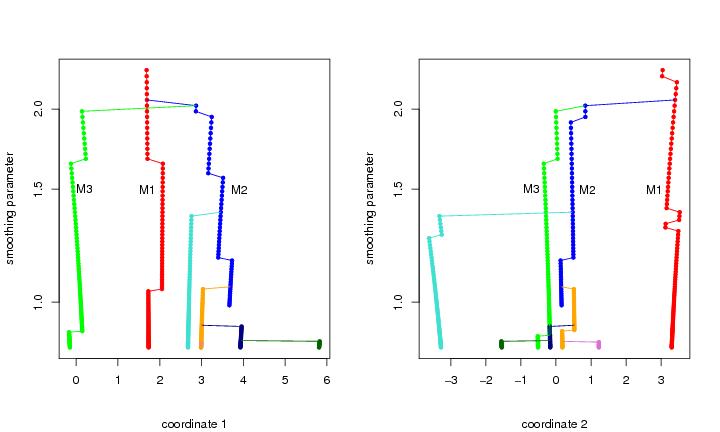
dendat<-sim.data(n=200,seed=5,type="mulmod")
h1<-0.85
h2<-2.3
lkm<-100
base<-10
hseq<-hgrid(h1,h2,lkm,base)
N<-c(30,30)
# Warning: the calculation can take a long time
estiseq<-lstseq.kern(dendat,hseq,N,lstree=TRUE,level=0.1,kernel="epane")
mt<-modegraph(estiseq)
# frame 1
plotmodet(mt,coordi=1,ylab="smoothing parameter")
text(0.2,1.5,"M3")
text(1.7,1.5,"M1")
text(3.9,1.5,"M2")
# frame 2
plotmodet(mt,coordi=2,ylab="smoothing parameter")
text(-0.7,1.5,"M3")
text(2.8,1.5,"M1")
text(0.9,1.5,"M2")
Figure 3

dendat<-sim.data(n=200,seed=5,type="mulmod")
h1<-0.85
h2<-2.3
lkm<-100
base<-10
hseq<-hgrid(h1,h2,lkm,base)
N<-c(30,30)
# Warning: the calculation can take a long time
estiseq<-lstseq.kern(dendat,hseq,N,lstree=TRUE,level=0.1,kernel="epane")
ii<-2
h<-estiseq$hseq[hind[ii]]
pcf<-estiseq$pcfseq[[hind[ii]]]
lst<-estiseq$lstseq[[hind[ii]]]
td<-treedisc(lst,pcf,30)
fb<-findbranch(td$parent,colo="blue",pch=24)
em<-excmas(td)
em<-em/em[1]
emr<-matrix("",length(em),1)
for (i in 1:length(fb$indicator)){
if (fb$indicator[i]==1){
emr[i]<-round(10000*em[i])/10000
fb$colovec[td$parent[i]]<-"red1"
fb$pchvec[td$parent[i]]<-22
}
}
bm<-branchmap(estiseq)
hindeksi<-101-hind[ii]
x<-bm$level
y<-bm$z[,hindeksi]
alku<-(dim(bm$z)[1]-1)*hindeksi+2
loppu<-alku+dim(bm$z)[1]-1
col<-bm$col[alku:loppu]
col[length(col)]<-"white"
col[length(col)-1]<-"white"
node1<-27
node2<-39
node3<-57
# frame 1
plottree(td,ptext=0.003,info=emr,xmarginright=0.25,xmarginleft=0.05,
infopos=-0.3,infolift=0.0005,col=fb$colovec,pch=fb$pchvec,cex=1.5,nodemag=2,
cex.axis=1.5,ylab="level",cex.lab=1.5)
# frame 2
plot(x,y,col=col,lty=1,xlab="level",ylab="excess mass",pch=19,
cex.lab=1.5,cex.axis=1.5)
segments(x[node1+1],y[node1],x[node1+1],y[node1+1],lty=1)
segments(x[node2+1],y[node2],x[node2+1],y[node2+1])
segments(x[node3+1],y[node3],x[node3+1],0)
Figure 4
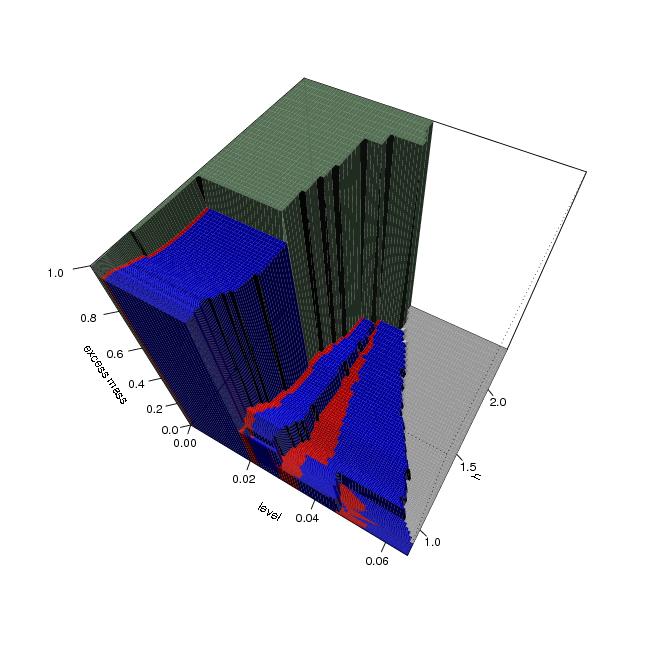
dendat<-sim.data(n=200,seed=5,type="mulmod")
h1<-0.85
h2<-2.3
lkm<-100
base<-10
hseq<-hgrid(h1,h2,lkm,base)
N<-c(30,30)
# Warning: the calculation can take a long time
estiseq<-lstseq.kern(dendat,hseq,N,lstree=TRUE,level=0.1,kernel="epane")
bm<-branchmap(estiseq)
# frame
plotbranchmap(bm)
